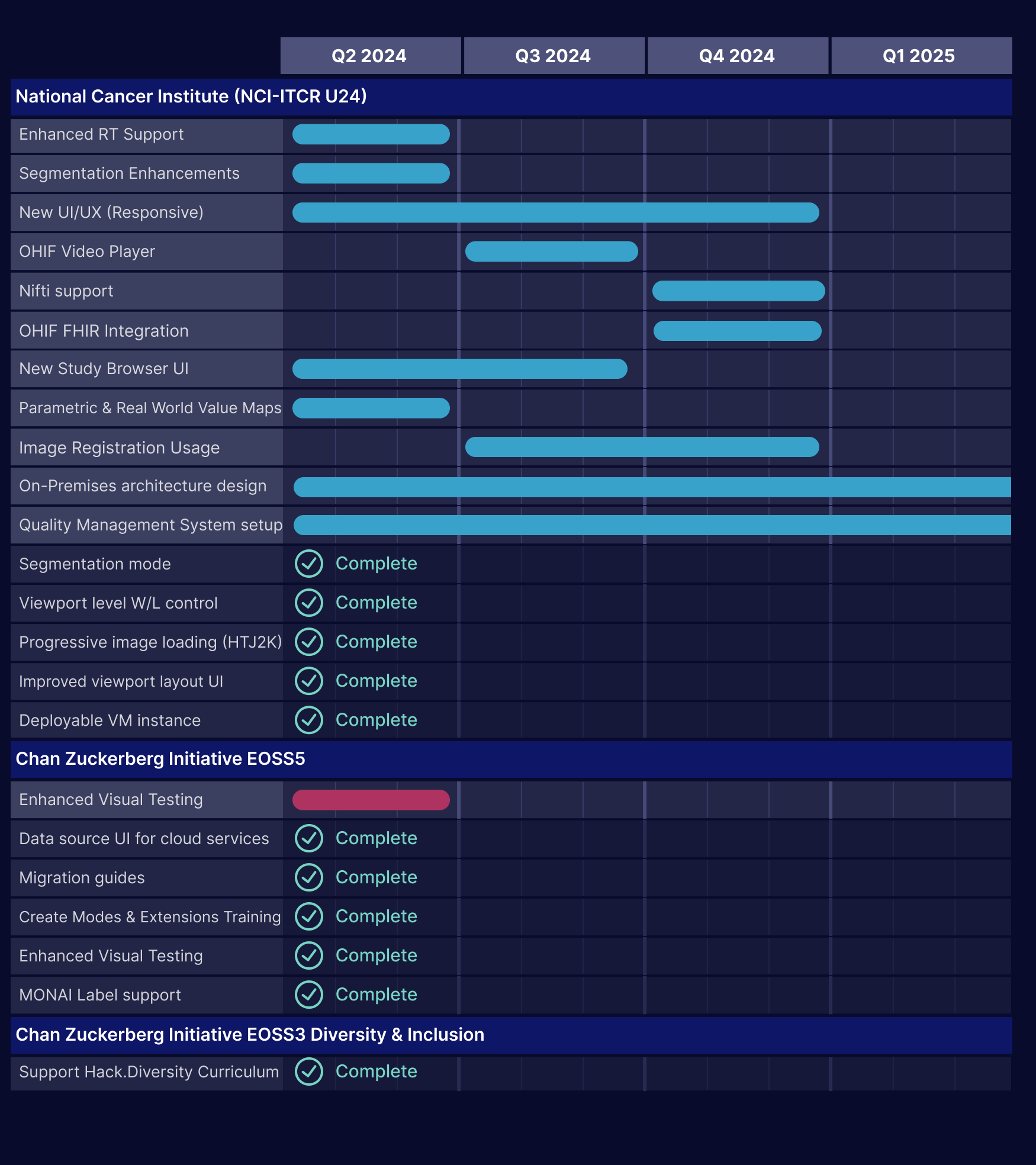
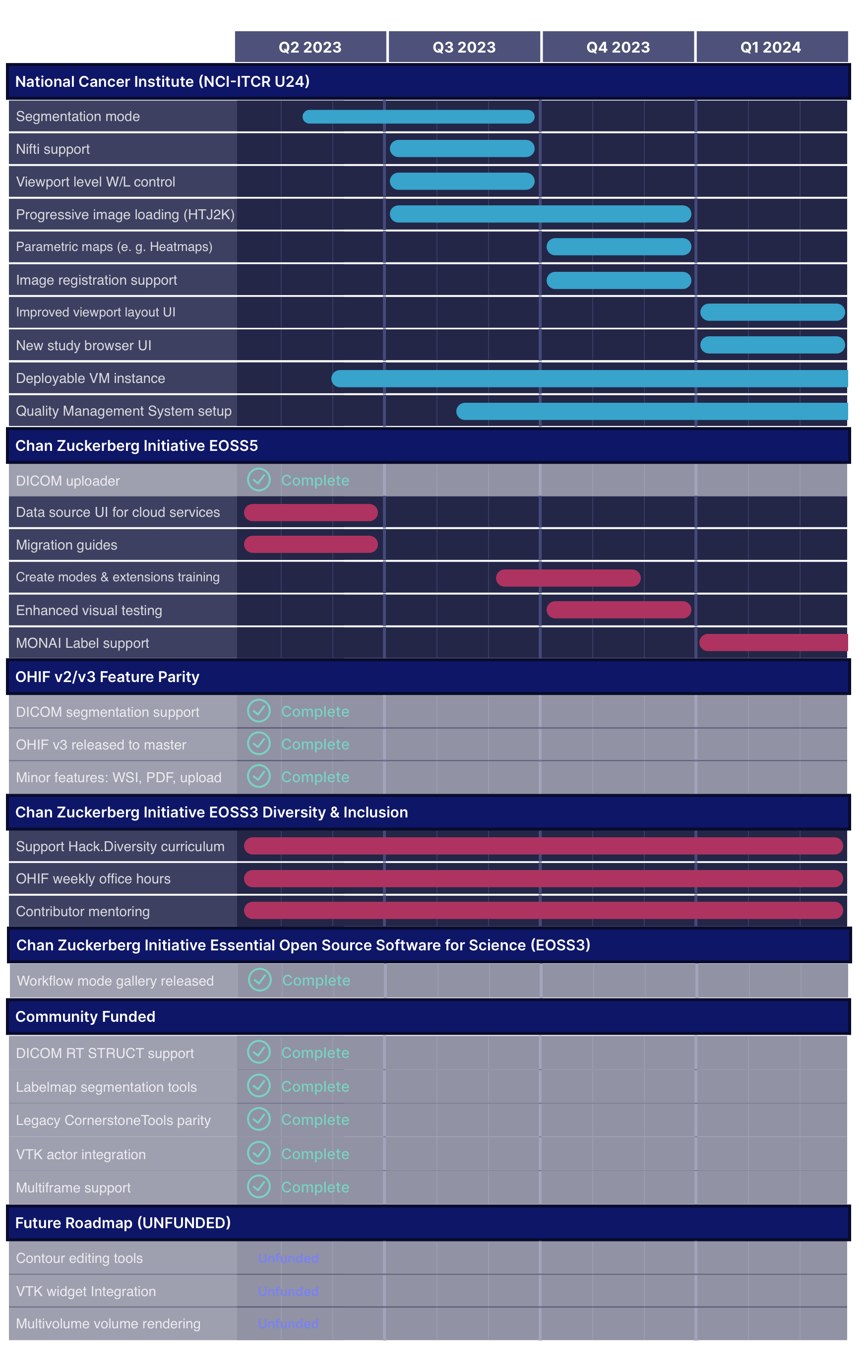
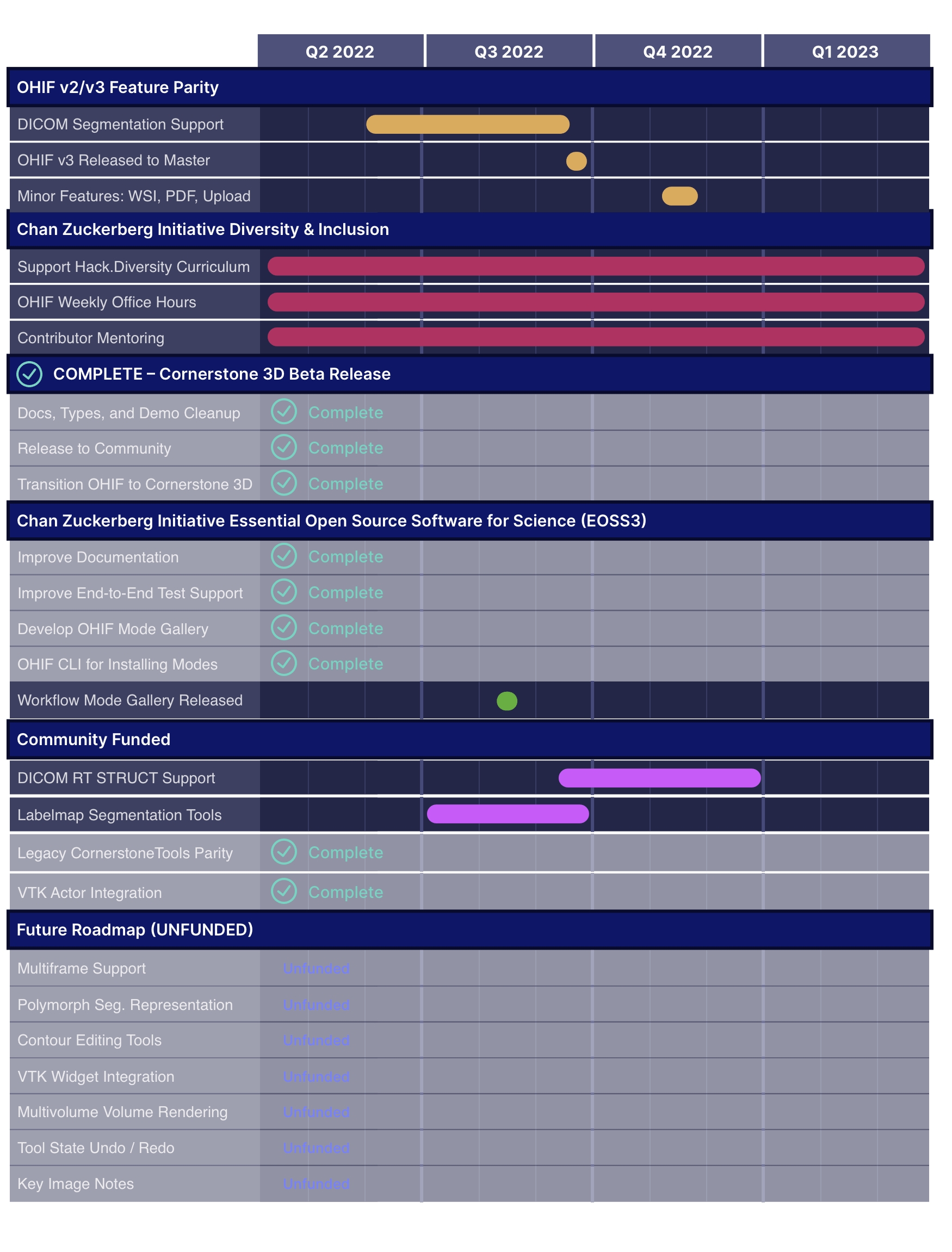
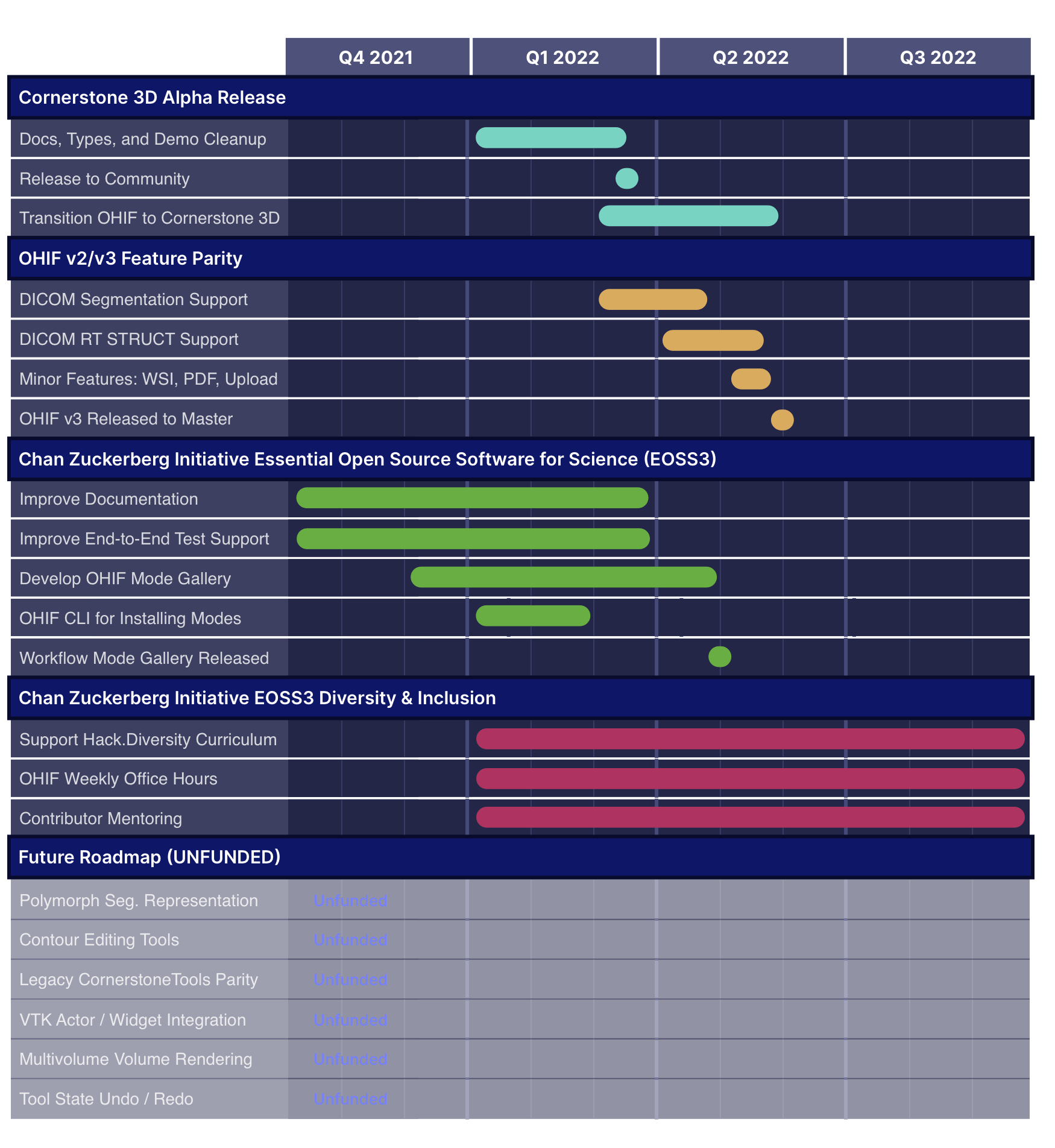
Archived Roadmaps
See the latest roadmaps here

With the viewer page now migrated to ohif/ui-next in version 3.10, the Worklist is the last major component still relying on the legacy UI. We're planning to migrate and redesign it with responsive layouts, better data handling, and support for thumbnails and series selection. This RFC is open for feedback.

We will introduce support for Radiation Therapy (RT) Dose visualization within the OHIF Viewer. Users will be able to overlay RT Dose data directly onto imaging modalities (e.g., CT, MR), enabling improved assessment and planning in radiation therapy workflows.

We will enhance OHIF Viewer with advanced multi-planar visualization for Radiation Therapy Structure Sets (RTSS). Users will be able to reconstruct and accurately visualize structure contours in arbitrary planes beyond the original acquisition plane, significantly improving precision and flexibility in radiation treatment planning.

We're expanding the viewport dialog to support advanced data fusion capabilities. Users will easily overlay and manage multiple datasets (e.g., PET on CT, RT Dose overlays) directly within each viewport, with intuitive controls for opacity, visibility, and blending.

We are finalizing our UI/UX overhaul by migrating all remaining components to the new ohif/ui-next. This update will fully replace the outdated OHIF/UI, offering enhanced customization, accessibility, and compatibility. Migration guides will be provided to ensure a smooth transition.

We will introduce undo/redo capabilities for annotation and segmentation actions, allowing users to reverse or reapply changes step-by-step. This feature will support configurable undo stack depth, with programmatic control to enable, disable, or clear the undo/redo history as needed.

Introducing a 3D brush tool for voxel-level segmentation, this tool enables selective region filling based on grayscale similarity and spatial connectivity. Users benefit from interactive previews, adjustable parameters, and integration with existing segmentations for precise, dynamic 3D editing.

We will develop methods that enable retrieval of detailed segmentation metrics, including intensity values, metabolic tumor volume, lesion glycolysis, and other critical statistics. This will support both individual labelmap segmentations and groups of overlapping segmentations, facilitating advanced analysis.

We will add functionality to interpolate label map segmentations across slices, providing users with a smooth, continuous segmentation experience. This tool will include a dedicated UI and will leverage algorithms from ITK to support interpolation between non-adjacent labelmap slices.

We will add a WebGPU-accelerated Grow Cut segmentation tool to our library, enabling fast, 3D segmentation. This tool will offer both manual and automatic modes, allowing users to configure initial regions for precise or one-click segmentation.

We will leverage AI models to enable automatic propagation and adjustment of segments across image slices. This functionality will use encoder-decoder models such as Segment Anything Model (SAM), allowing users to efficiently extend and adapt segmentations slice by slice.

We will improve the rendering of segmentation outlines in MIP viewports, ensuring that outlines accurately follow the anatomy from all viewing angles. This enhancement will provide configurable outline thickness and color, as well as interactive selection of segments.

We will create a tool that identifies and places bidimensional measurements on the largest in-plane slice of a 3D segment. This includes the longest diameter along the lesion’s main axis and the longest possible perpendicular diameter within the same slice.

We will add functionality to highlight individual segments on hover by increasing their border thickness, making it easier for users to identify and interact with specific lesions. This enhancement will require adjustments to the WebGL shader for dynamic border control.






















Archived Roadmaps
See the latest roadmaps here

With the viewer page now migrated to ohif/ui-next in version 3.10, the Worklist is the last major component still relying on the legacy UI. We're planning to migrate and redesign it with responsive layouts, better data handling, and support for thumbnails and series selection. This RFC is open for feedback.

We will introduce support for Radiation Therapy (RT) Dose visualization within the OHIF Viewer. Users will be able to overlay RT Dose data directly onto imaging modalities (e.g., CT, MR), enabling improved assessment and planning in radiation therapy workflows.

We will enhance OHIF Viewer with advanced multi-planar visualization for Radiation Therapy Structure Sets (RTSS). Users will be able to reconstruct and accurately visualize structure contours in arbitrary planes beyond the original acquisition plane, significantly improving precision and flexibility in radiation treatment planning.

We're expanding the viewport dialog to support advanced data fusion capabilities. Users will easily overlay and manage multiple datasets (e.g., PET on CT, RT Dose overlays) directly within each viewport, with intuitive controls for opacity, visibility, and blending.

We are finalizing our UI/UX overhaul by migrating all remaining components to the new ohif/ui-next. This update will fully replace the outdated OHIF/UI, offering enhanced customization, accessibility, and compatibility. Migration guides will be provided to ensure a smooth transition.

We will introduce undo/redo capabilities for annotation and segmentation actions, allowing users to reverse or reapply changes step-by-step. This feature will support configurable undo stack depth, with programmatic control to enable, disable, or clear the undo/redo history as needed.

Introducing a 3D brush tool for voxel-level segmentation, this tool enables selective region filling based on grayscale similarity and spatial connectivity. Users benefit from interactive previews, adjustable parameters, and integration with existing segmentations for precise, dynamic 3D editing.

We will develop methods that enable retrieval of detailed segmentation metrics, including intensity values, metabolic tumor volume, lesion glycolysis, and other critical statistics. This will support both individual labelmap segmentations and groups of overlapping segmentations, facilitating advanced analysis.

We will add functionality to interpolate label map segmentations across slices, providing users with a smooth, continuous segmentation experience. This tool will include a dedicated UI and will leverage algorithms from ITK to support interpolation between non-adjacent labelmap slices.

We will add a WebGPU-accelerated Grow Cut segmentation tool to our library, enabling fast, 3D segmentation. This tool will offer both manual and automatic modes, allowing users to configure initial regions for precise or one-click segmentation.

We will leverage AI models to enable automatic propagation and adjustment of segments across image slices. This functionality will use encoder-decoder models such as Segment Anything Model (SAM), allowing users to efficiently extend and adapt segmentations slice by slice.

We will improve the rendering of segmentation outlines in MIP viewports, ensuring that outlines accurately follow the anatomy from all viewing angles. This enhancement will provide configurable outline thickness and color, as well as interactive selection of segments.

We will create a tool that identifies and places bidimensional measurements on the largest in-plane slice of a 3D segment. This includes the longest diameter along the lesion’s main axis and the longest possible perpendicular diameter within the same slice.

We will add functionality to highlight individual segments on hover by increasing their border thickness, making it easier for users to identify and interact with specific lesions. This enhancement will require adjustments to the WebGL shader for dynamic border control.

























